
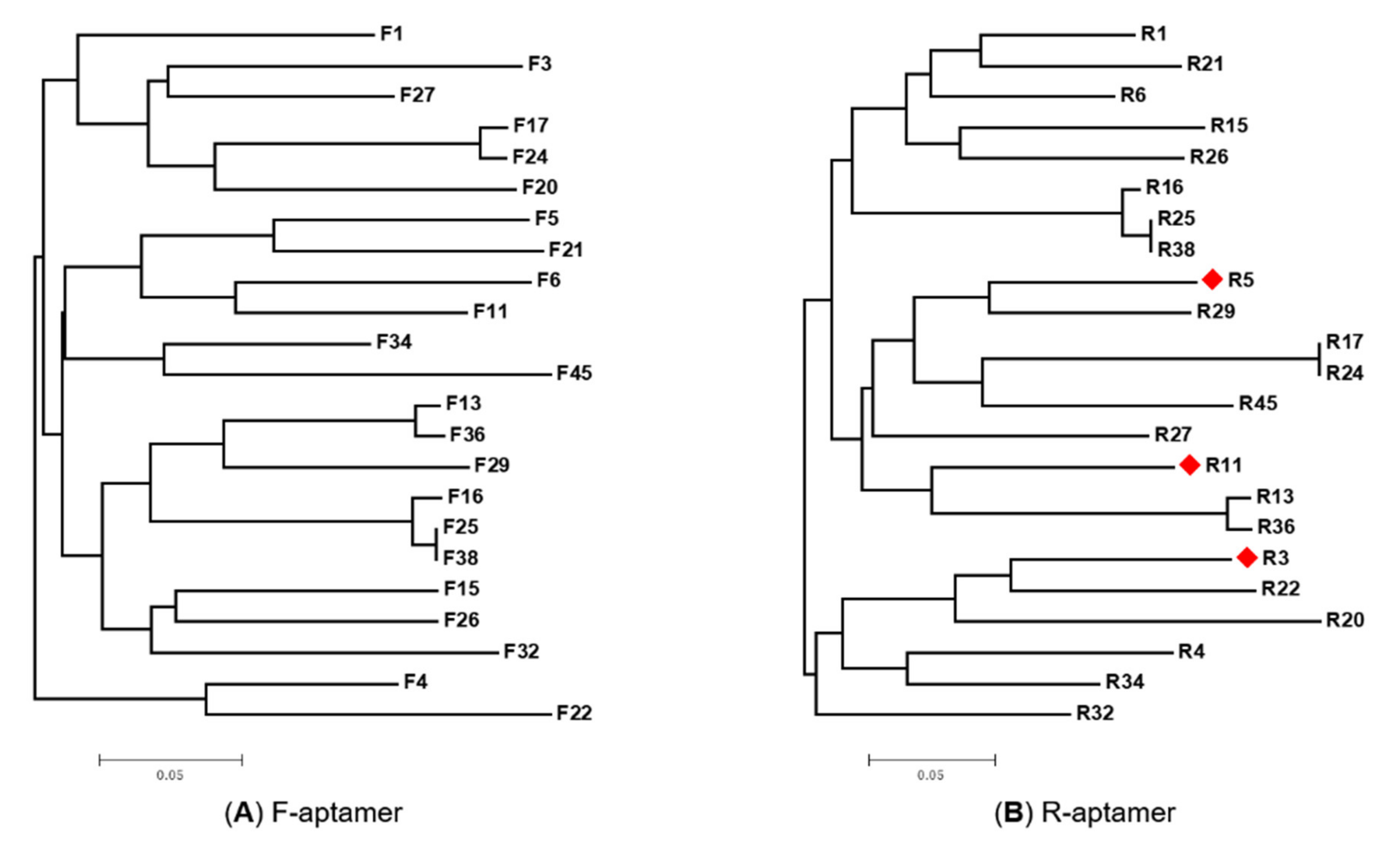
2019 Sherlock and Breaker 2020 Husser et al. In general, interaction with macromolecules (e.g., proteins or nucleic acids) does not necessarily involve high structuralĬomplexity, on the contrary, the tight and specific recognition of small ligands usually occurs through sophisticated structuresīest exemplified by those found in riboswitches aptamer domains ( Winkler and Breaker 2005 Pavlova et al. To the three-dimensional architecture of the molecule and its capacity to properly display key residues in space. RNA is able to perform a wide range of functions (e.g., scaffolding, recognition, or catalysis) that are intimately linked Previous Section Next Section INTRODUCTION With an up to 10-fold improved performance. Only shed some new light on the way this aptamer can be engineered, but it also allowed us to easily identify new variants We demonstrate the efficiency of this “µIVC-Useq” technologyīy rapidly identifying a set of sequences readily accepted by a key domain of the light-up RNA aptamer SRB-2.
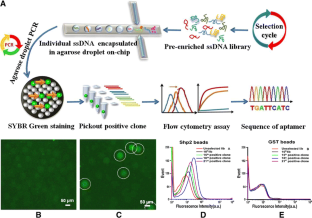
Neural network enabling unsupervised bioinformatic analysis. In the present work, we further increased the automation level of the pipeline by implementing an artificial We later improved µIVC efficiencyīy using it in tandem with high-throughput sequencing, though a laborious bioinformatic step was still required at the end In which reactions are performed in picoliter droplets at rates of several thousand per second. We introduced the microfluidic-assisted in vitro compartmentalization (µIVC), an efficient and cost-effective screening strategy That may not only help in validating the model, but also inform on the mutational robustness of a structural element and theĮxtent to which a sequence can be modified without altering RNA function, an important set of information to assist RNA engineering. Such limitations can be overcome using high-throughput functional screenings Which can rapidly become time consuming and laborious. Yet these models still require experimental validation through the preparation and functional assay of mutants, Probing, or in silico predictions have been developed to establish structural models, sometimes with a precision down to atomic Many approaches encompassing X-ray crystallography, NMR, structural The function of an RNA is intimately linked to its structure.


 0 kommentar(er)
0 kommentar(er)
